| A method to compare costs at given error
rates for proposed sequencing technologies |
|
|
| Human
diploid genome goal = $1K/6e6 kb @
error<1e-8 = |
1.7E-4 |
$/kb |
|
| (exact
size of gaps as yet unspecified but expected similar to current) |
|
|
|
|
| Coverage |
SBL Err |
SBL $/kb |
SBL2 $/kb |
ABI Err |
ABI $/kb |
454 Err |
454 $/kb |
|
| 1 |
3E-3 |
0.09 |
1.2E-4 |
3E-4 |
0.90 |
4E-2 |
0.19 |
|
| 6.5 |
3E-7 |
0.59 |
7.8E-4 |
|
|
|
|
|
| 10 |
|
|
1E-6 |
9.00 |
|
|
|
| 40 |
|
|
|
|
|
4E-5 |
7.68 |
|
|
|
|
|
|
|
|
|
SBL-estimated |
|
ABI-estimated |
|
454-estimated |
|
| Errors |
Coverage |
SBL $/kb |
SBL2 $/kb |
Coverage |
ABI $/kb |
Coverage |
454 $/kb |
|
| 1E-2 |
|
|
|
|
9 |
1.7 |
|
| 1E-3 |
1.7 |
0.15 |
2.0E-4 |
|
22 |
4.2 |
|
| 1E-4 |
3.0 |
0.27 |
3.6E-4 |
2.7 |
2.5 |
35 |
6.7 |
|
| 1E-5 |
4.4 |
0.40 |
5.3E-4 |
6.4 |
5.7 |
48 |
9.2 |
|
| 1E-6 |
5.8 |
0.52 |
6.9E-4 |
10.0 |
9.0 |
61 |
11.7 |
|
| 1E-7 |
7.2 |
0.64 |
8.6E-4 |
13.6 |
12.3 |
74 |
14.2 |
|
| 1E-8 |
8.5 |
0.77 |
1.0E-3 |
17.3 |
15.5 |
87 |
16.7 |
|
|
|
|
|
| SBL
cost and error rates from Shendure et al Science 2005 |
|
| ABI
cost and error estimates from Broad Inst. & Agencourt |
|
| 454
cost estimate: 26.7 Mbp/($5K
reagents+$500K/(3*365*4) 3yr_equip
amortization 4 runs/day) |
|
| 454
error rates from Table 2 of Margulies et al Nature 2005 |
|
| Estimated
required coverage = c1+(c1-c2)*[log(e)-log(e1)] / [log(e1)-log(e2)] |
|
| The
above formula is inapplicable once the error rate plateaus into
"systematic" (non-random) errors |
|
| SBL2
scenario: as SBL except 4 CCD pixel per bp on each cycle instead of current
750, |
|
| plus lower fractional cost of library for
human, simple savings on reagents, and dual flow cell. |
|
|
|
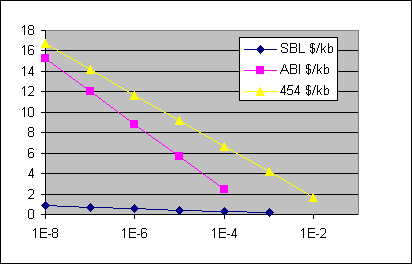
| Errors |
SBL $/kb |
ABI $/kb |
454 $/kb |
| 1.E-02 |
|
1.7 |
| 1.E-03 |
0.2 |
|
4.2 |
| 1.E-04 |
0.3 |
2.4 |
6.7 |
| 1.E-05 |
0.4 |
5.6 |
9.2 |
| 1.E-06 |
0.6 |
8.9 |
11.7 |
| 1.E-07 |
0.7 |
12.1 |
14.2 |
| 1.E-08 |
0.9 |
15.3 |
16.7 |
|
|
|
|
|
|
|
|
|
|
|
|