




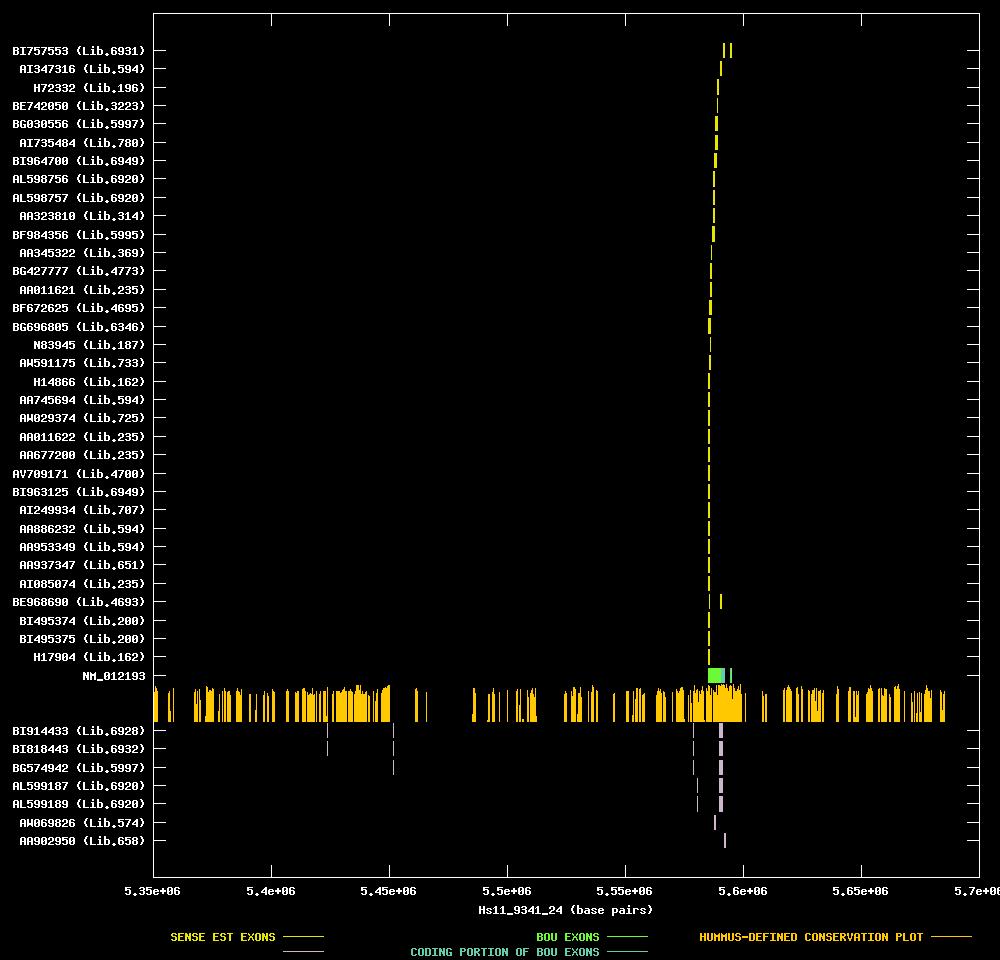
| Candidate UniGene cluster: | UniGene Cluster Hs.19545 |
| Description: | frizzled (Drosophila) homolog 4 |
| Best-Of-UniGene (BOU) Sequence | NM_012193 |
| Genomic Coordiantes Displayed: | Bases 5350000 to 5700000 of contig Hs11_9341_24 |
| BOU Orientation Along Contig: | RIGHT-TO-LEFT with respect to contig |
| Link to JPEG of genomic mapping | Hs.19545.jpeg |
| Best sense EST/protein match: | BI757553 matched ref|NP_036325.1| (NM_012193) frizzled homolog 4 (Drosophila); frizzled (Drosophila) (E = e-124) |
| Best antisense EST/protein match: | No protein match with an e-value of less than 1e-10 |
ANTISENSE ESTs
| BI914433 | cDNA clone IMAGE:5246259 | brain | 5' read |  | ||
| BI818443 | cDNA clone IMAGE:5174092 | pooled brain, lung, testis | 5' read |  | ||
| BG574942 | cDNA clone IMAGE:4706743 | mammary adenocarcinoma, cell line | 5' read |  | ||
| AL599187 | cDNA clone DKFZp313P1924 | 5' read | ||||
| AL599189 | cDNA clone DKFZp313P2024 | 5' read |  | |||
| AW069826 | cDNA clone HBMSC_cr52b03 | bone | 3' read | |||
| AA902950 | cDNA clone IMAGE:1516769 | smooth muscle | 3' read | 0.6 kb |  |
| H17904 | cDNA clone IMAGE:50620 | brain | 3' read | 0.6 kb |  | |
| BI495375 | cDNA clone IMAGE:2539916 | ear | 5' read |  | ||
| BI495374 | cDNA clone IMAGE:2539916 | ear | 3' read |  | ||
| BE968690 | cDNA clone IMAGE:3933940 | heart | 5' read | |||
| AI085074 | cDNA clone IMAGE:1653956 | pool | 3' read | 1.3 kb |  | |
| AA937347 | cDNA clone IMAGE:1491534 | skin | 3' read | 1.0 kb |  | |
| AA953349 | cDNA clone IMAGE:1573258 | kidney | 3' read | 0.7 kb |  | |
| AA886232 | cDNA clone IMAGE:1492849 | kidney | 3' read | 0.7 kb |  | |
| AI249934 | cDNA clone IMAGE:2000841 | lymph | 3' read | 1.9 kb |  | |
| BI963125 | cDNA clone (no-name) | islets of langerhans | 3' read |  | ||
| AV709171 | cDNA clone ADCAVG12 | adrenal gland | 5' read |  | ||
| AA677200 | cDNA clone IMAGE:454698 | pool | 3' read |  | ||
| AA011622 | cDNA clone IMAGE:429599 | pool | 3' read | 1.3 kb |  | |
| AW029374 | cDNA clone IMAGE:2543194 | stomach | 3' read | 0.7 kb |  | |
| AA745694 | cDNA clone IMAGE:1323968 | kidney | 3' read | 0.7 kb |  | |
| H14866 | cDNA clone IMAGE:49122 | brain | 5' read | 0.6 kb | ||
| AW591175 | cDNA clone IMAGE:2703691 | uterus | 3' read | |||
| N83945 | cDNA clone KK4065 | heart | 5' read |