
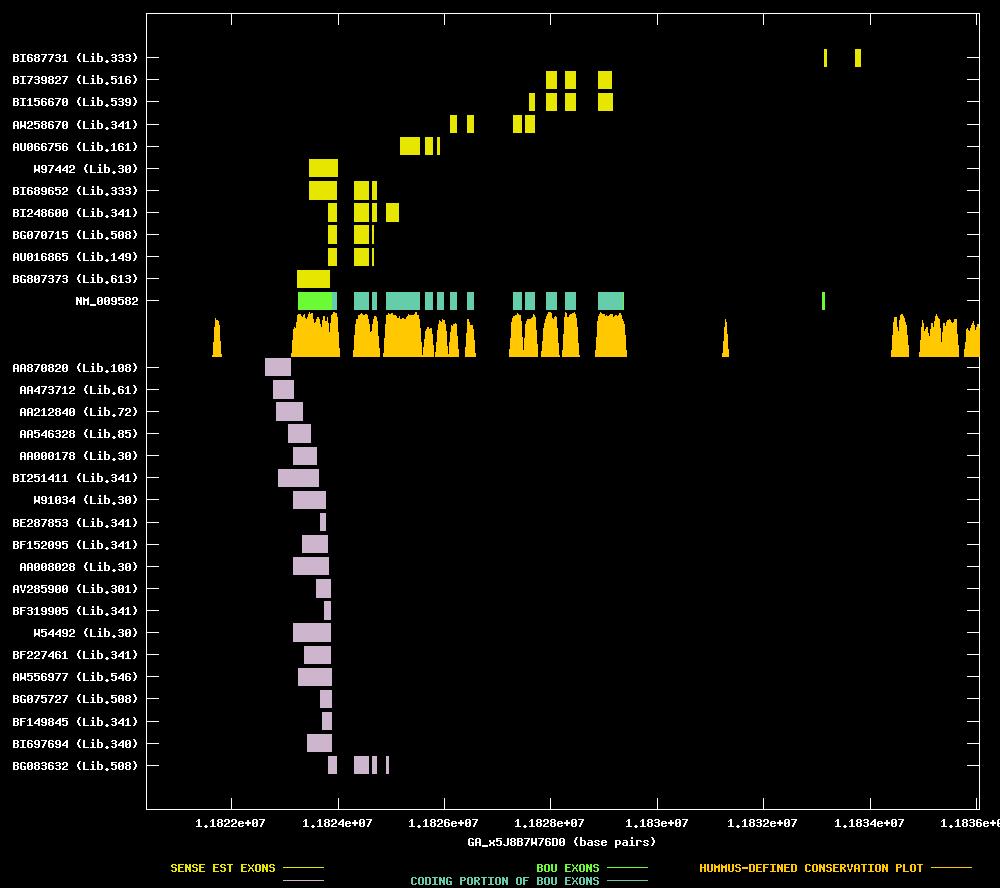
| Candidate UniGene cluster: | UniGene Cluster Mm.4358 |
| Description: | mitogen activated protein kinase kinase kinase 12 |
| Best-Of-UniGene (BOU) Sequence | NM_009582 |
| Genomic Coordiantes Displayed: | Bases 11820397 to 11836058 of contig GA_x5J8B7W76D0 |
| BOU Orientation Along Contig: | RIGHT-TO-LEFT with respect to contig |
| Link to JPEG of genomic mapping | Mm.4358.jpeg |
| Best sense EST/protein match: | AW258670 matched ref|NP_033608.1| (NM_009582) zipper (leucine) protein kinase [Mus musculus] (E = e-101) |
| Best antisense EST/protein match: | No protein match with an e-value of less than 1e-10 |
ANTISENSE ESTs
| AA870820 | cDNA clone IMAGE:1095417 | 5' read | ||||
| AA473712 | cDNA clone IMAGE:866157 | embryo | 5' read | |||
| AA212840 | cDNA clone IMAGE:677201 | liver | 5' read | |||
| AA546328 | cDNA clone IMAGE:945650 | embryo | 5' read | |||
| AA000178 | cDNA clone IMAGE:425631 | embryo | 5' read | |||
| BI251411 | cDNA clone IMAGE:5150264 | tumor, gross tissue | 5' read | |||
| W91034 | cDNA clone IMAGE:421154 | embryo | 5' read | |||
| BE287853 | cDNA clone IMAGE:3497296 | tumor, gross tissue | 5' read | |||
| BF152095 | cDNA clone IMAGE:3670064 | tumor, gross tissue | 5' read | |||
| AA008028 | cDNA clone IMAGE:438406 | embryo | 5' read | |||
| AV285900 | cDNA clone 5031415E16 | ovary and uterus | ||||
| BF319905 | cDNA clone IMAGE:3665728 | tumor, gross tissue | 3' read | |||
| W54492 | cDNA clone IMAGE:367895 | embryo | 5' read | |||
| BF227461 | cDNA clone IMAGE:3670064 | tumor, gross tissue | 3' read | |||
| AW556977 | cDNA clone L0275G08 | 3' read | ||||
| BG075727 | cDNA clone H3150H08 | 3' read | ||||
| BF149845 | cDNA clone IMAGE:3665728 | tumor, gross tissue | 5' read | |||
| BI697694 | cDNA clone IMAGE:5376475 | tumor, biopsy sample | 5' read | |||
| BG083632 | cDNA clone H3090D01 | 5' read |  |
| BG807373 | cDNA clone (no-name) | neural retina | ||||
| AU016865 | cDNA clone J0731A03 | embryo | 3' read |  | ||
| BG070715 | cDNA clone H3090D01 | 3' read |  | |||
| BI248600 | cDNA clone IMAGE:5148680 | tumor, gross tissue | 5' read |  | ||
| BI689652 | cDNA clone IMAGE:5356163 | infiltrating ductal carcinoma | 5' read |  | ||
| W97442 | cDNA clone IMAGE:422079 | embryo | 5' read |  | ||
| AU066756 | cDNA clone MNCb-2527 | brain | 5' read |  | ||
| AW258670 | cDNA clone IMAGE:2811677 | tumor, gross tissue | 5' read |  | ||
| BI156670 | cDNA clone IMAGE:5061643 | tumor, gross tissue | 5' read |  | ||
| BI739827 | cDNA clone IMAGE:5369193 | retina | 5' read |  | ||
| BI687731 | cDNA clone IMAGE:5355872 | infiltrating ductal carcinoma | 5' read |