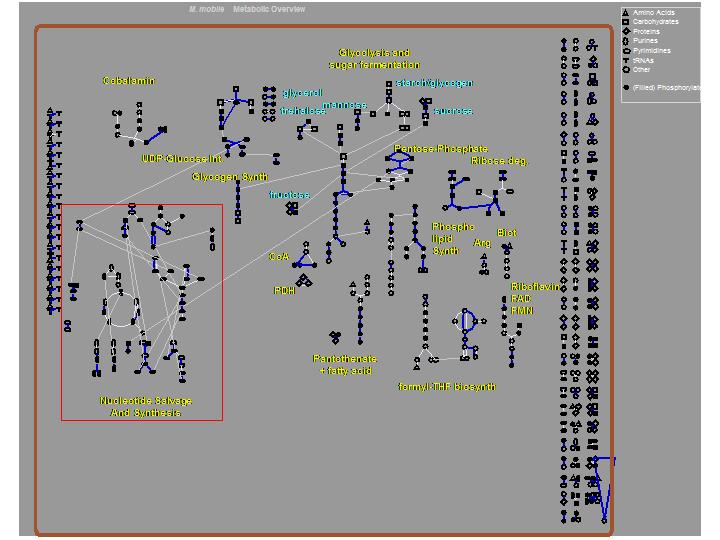
The M. mobile genome was completely sequenced and annotated as a collaboration between the Broad Institute and the Church Lab at Harvard Medical School. These efforts were completed in April 2004. This page is dedicated to the metabolic analysis of the M. mobile genome and proteome. We hope to make measurements of various metabolites in vivo and to identify novel protein components that carry out required functions in the cell. A preliminary metabolic model was generated directly from the annotated genome using the PathwayTools and MetaCyc software packages, and then manually curated by our research team. An overview of the model is shown below. General genome sequence and annotation information can be obtained from the Broad Institute's M. mobile genome homepage. We will continue to update this model and add content as more data are collected and incorporated into the model.

Key:
Dark blue connections=reactions present in M. mobile.
White connections=reactions absent in M. mobile, but part of the pathway represented.
Gray connections=connections between identical metabolites in different pathways.