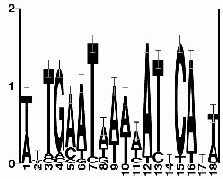
 sequence logo for
ArgR regulatory motif
sequence logo for
ArgR regulatory motif
|
www links: |
Abigail Manson McGuire, Jason D. Hughes, and George M. Church
Download full text of course document in Postscript or Microsoft Word formats.
AlignACE exercise
Short conserved sequence elements located upstream of the transcriptional start site are often binding sites for transcription factors that regulate a group of genes involved in a similar cellular function. Additional genes with this motif in their upstream regions are good candidates for involvement in the same cellular function. Thus, upstream regulatory motifs can provide powerful hypotheses about links in the genetic regulatory network. These upstream regulatory motifs can be discovered computationally by local alignment of upstream regions from coregulated sets of genes. Here we will focus on the AlignACE motif-finding algorithm (Roth et al., 1998), which is a modified version of the Gibbs sampler algorithm (Neuwald et al., 1995).
In order to discover a motif computationally using local alignment methods, an accurate list of coregulated genes, or regulon, is needed. The AlignACE program can tolerate some noise in the alignment process from extra sequence that does not contain the motif of interest, but too much extra sequence will prevent the motif from being aligned. Thus, we must employ additional experimental and computational methods to obtain accurate regulon predictions. mRNA expression data has proven useful for this task. Genes which are up-regulated or down-regulated in a single experimental condition are likely to be coregulated, as well as genes with similar mRNA expression profiles across many conditions or timepoints. There are also several computational methods for predicting regulons. Here we will discuss several methods for predicting regulons, motif-discovery algorithms, indices for evaluating the significance of a motif, and applications of motif-finding using AlignACE in 17 bacterial genomes and in Saccharomyces cerevisiae.
 sequence logo for
ArgR regulatory motif
sequence logo for
ArgR regulatory motif
|
www links: |
| Abigail Manson McGuire | |
| Harvard Medical School Genetics Dept., Church Lab Alpert Bldg. 512 200 Longwood Ave. Boston, MA. 02115 | Phone: (617) 432-0503 Fax: (617) 432-7266 |